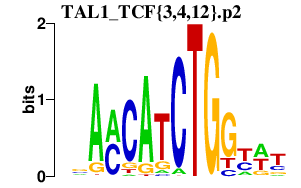
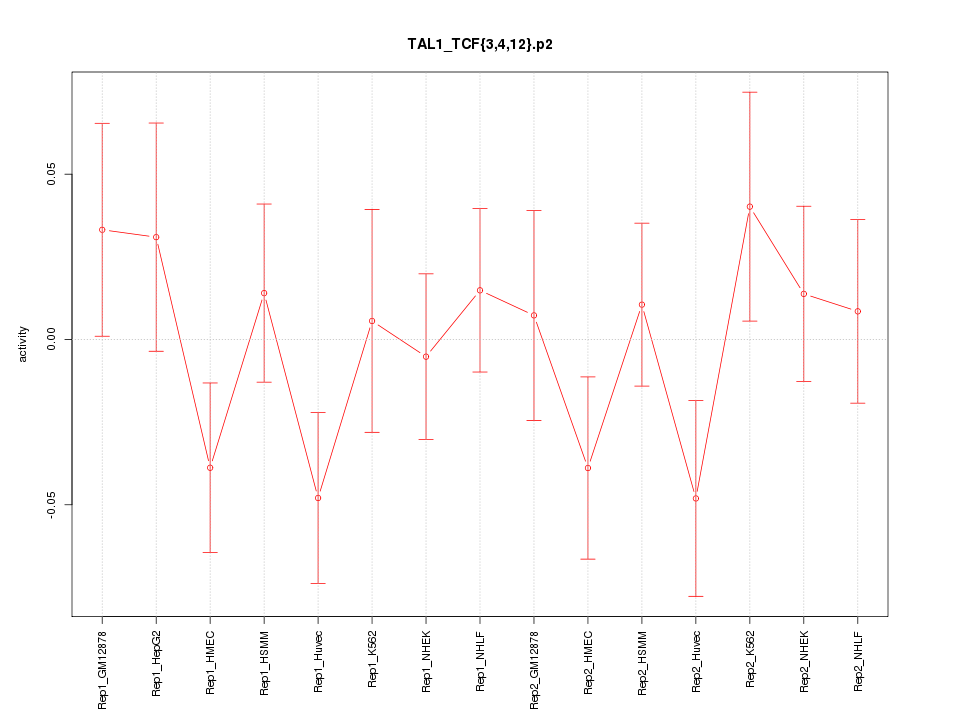
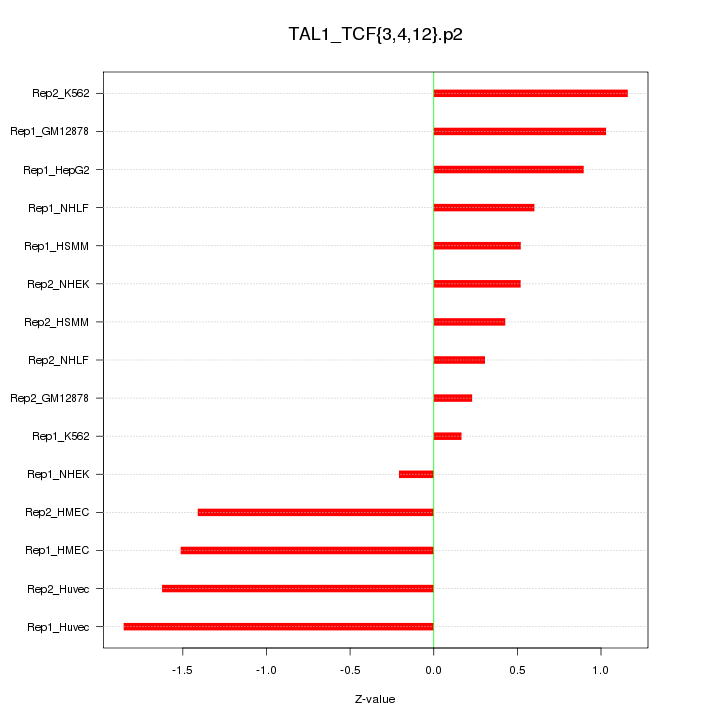
|
chr12_-_46405477
|
2.388
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr22_+_38652555
|
1.635
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr17_+_38256886
|
1.521
|
|
|
|
|
chr19_+_17499064
|
1.290
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr17_+_38256710
|
1.280
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr16_+_56286212
|
1.280
|
NM_032269
|
CCDC135
|
coiled-coil domain containing 135
|
|
chr1_+_179164195
|
1.235
|
|
|
|
|
chr10_-_73518772
|
1.224
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr10_+_85923473
|
1.069
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr17_+_45617676
|
1.036
|
|
|
|
|
chr1_+_221168388
|
1.020
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr2_+_191009265
|
0.979
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr11_-_5212168
|
0.952
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr1_-_205272619
|
0.926
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr2_+_169367345
|
0.900
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chrX_-_154216929
|
0.876
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr17_-_53761119
|
0.795
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr16_+_11966464
|
0.760
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr4_-_185921384
|
0.748
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr11_+_46651090
|
0.730
|
|
ATG13
|
ATG13 autophagy related 13 homolog (S. cerevisiae)
|
|
chr3_+_134976965
|
0.729
|
|
TF
|
transferrin
|
|
chr6_-_30188845
|
0.724
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr12_+_115481568
|
0.692
|
NM_001085481
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2
|
|
chr22_+_23309717
|
0.690
|
NM_013430
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr2_-_25320299
|
0.662
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr14_-_93493519
|
0.661
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr12_-_101835183
|
0.626
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr5_+_53787187
|
0.625
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr2_-_157892391
|
0.617
|
NM_001009959
|
ERMN
|
ermin, ERM-like protein
|
|
chr1_-_144426918
|
0.614
|
NM_007053
|
CD160
|
CD160 molecule
|
|
chr1_+_19807168
|
0.590
|
|
|
|
|
chr20_-_39199936
|
0.582
|
|
|
|
|
chr22_+_22907443
|
0.572
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr17_-_43390108
|
0.571
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr8_-_68821131
|
0.569
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr19_+_4442610
|
0.566
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr17_-_9863929
|
0.557
|
|
GAS7
|
growth arrest-specific 7
|
|
chr1_-_169888373
|
0.555
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chrX_+_46825179
|
0.547
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr16_+_4546491
|
0.545
|
NM_001145011
|
LOC342346
|
hypothetical protein LOC342346
|
|
chr2_-_158008763
|
0.536
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr12_-_101835510
|
0.519
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr11_+_5367182
|
0.515
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr19_+_61624
|
0.504
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr10_+_80743769
|
0.499
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr3_+_120982020
|
0.498
|
NM_003889
NM_033013
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr1_-_224143389
|
0.487
|
NM_020997
|
LEFTY1
|
left-right determination factor 1
|
|
chr22_-_34930705
|
0.486
|
NM_030643
NM_145660
|
APOL4
|
apolipoprotein L, 4
|
|
chr7_+_94953148
|
0.486
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr21_-_33107813
|
0.474
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_+_207926172
|
0.474
|
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr22_+_22907417
|
0.474
|
|
SUSD2
|
sushi domain containing 2
|
|
chr20_-_44612619
|
0.471
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr8_-_81155564
|
0.468
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr1_-_176205672
|
0.460
|
NM_033127
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr1_-_158948176
|
0.455
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr7_-_92693716
|
0.452
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr12_-_101835026
|
0.433
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr12_-_52016270
|
0.431
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr19_+_58239802
|
0.427
|
NM_001191055
|
HERV-V2
|
envelope glycoprotein ENVV2
|
|
chr2_-_183611470
|
0.426
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr5_-_134810696
|
0.425
|
NM_130848
|
C5orf20
|
chromosome 5 open reading frame 20
|
|
chr6_+_167456230
|
0.423
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr2_-_55131157
|
0.421
|
|
RTN4
|
reticulon 4
|
|
chr6_-_66473790
|
0.413
|
NM_001142800
NM_001142801
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr19_+_15699833
|
0.410
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr1_-_24179326
|
0.400
|
|
SRSF10
|
serine/arginine-rich splicing factor 10
|
|
chr1_+_54786394
|
0.394
|
NM_015547
NM_147161
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr5_-_131907071
|
0.386
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr1_-_199613427
|
0.382
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr16_-_79811475
|
0.380
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr17_+_53769562
|
0.377
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr2_-_133256097
|
0.376
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr21_+_17841297
|
0.374
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr2_-_55131124
|
0.372
|
|
RTN4
|
reticulon 4
|
|
chr9_+_95886625
|
0.370
|
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr1_-_203657803
|
0.367
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr9_+_114376709
|
0.363
|
|
KIAA1958
|
KIAA1958
|
|
chr7_-_36992221
|
0.358
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr2_+_46560207
|
0.357
|
NM_001145051
|
LOC388946
|
transmembrane protein ENSP00000343375
|
|
chrX_-_111809929
|
0.354
|
NM_178175
|
LHFPL1
|
lipoma HMGIC fusion partner-like 1
|
|
chr17_-_9864048
|
0.353
|
|
GAS7
|
growth arrest-specific 7
|
|
chr9_+_6318348
|
0.351
|
NM_001001874
NM_001001875
NM_033516
|
TPD52L3
|
tumor protein D52-like 3
|
|
chr2_-_151854619
|
0.351
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr2_-_55131176
|
0.349
|
|
RTN4
|
reticulon 4
|
|
chr12_-_6615348
|
0.349
|
NM_020400
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr4_+_14985272
|
0.343
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr15_+_56511466
|
0.334
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr11_-_68505030
|
0.334
|
NM_198923
|
MRGPRD
|
MAS-related GPR, member D
|
|
chr14_-_93919305
|
0.333
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr6_+_52159143
|
0.332
|
NM_002190
|
IL17A
|
interleukin 17A
|
|
chr8_+_21967008
|
0.326
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr3_+_73193499
|
0.326
|
NM_018029
|
EBLN2
|
endogenous Borna-like N element-2
|
|
chr10_+_11246934
|
0.323
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr16_-_67943157
|
0.312
|
NM_144676
|
TMED6
|
transmembrane emp24 protein transport domain containing 6
|
|
chr5_-_88235624
|
0.307
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr9_+_95886558
|
0.307
|
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr3_-_10307408
|
0.304
|
NM_001134941
NM_016362
|
GHRL
|
ghrelin/obestatin prepropeptide
|
|
chr10_+_44726635
|
0.303
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr12_+_56114724
|
0.301
|
NM_005538
|
INHBC
|
inhibin, beta C
|
|
chr7_+_129807467
|
0.301
|
NM_001868
|
CPA1
|
carboxypeptidase A1 (pancreatic)
|
|
chr10_+_5228756
|
0.301
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr19_+_40224480
|
0.298
|
|
HPN
|
hepsin
|
|
chr16_+_31105579
|
0.297
|
|
|
|
|
chr11_+_66979393
|
0.295
|
NM_145200
|
CABP4
|
calcium binding protein 4
|
|
chr22_+_34979118
|
0.285
|
|
APOL1
|
apolipoprotein L, 1
|
|
chr1_-_53972464
|
0.284
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr17_-_64568641
|
0.277
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr7_+_138695236
|
0.276
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr19_-_44060729
|
0.274
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr5_+_155686750
|
0.272
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr1_+_205328834
|
0.271
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr17_-_59342319
|
0.271
|
NM_001318
NM_022579
NM_022580
NM_022581
|
CSHL1
|
chorionic somatomammotropin hormone-like 1
|
|
chr3_+_23904086
|
0.270
|
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast)
|
|
chr2_-_188086612
|
0.270
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr17_+_72827227
|
0.270
|
|
SEPT9
|
septin 9
|
|
chr7_+_113853790
|
0.269
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr3_-_115960807
|
0.267
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_+_36318922
|
0.266
|
NM_173676
NM_001145716
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr12_-_53064723
|
0.263
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr2_-_55131096
|
0.258
|
|
RTN4
|
reticulon 4
|
|
chr20_-_42457717
|
0.252
|
|
|
|
|
chr6_-_127822227
|
0.252
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr22_+_20880112
|
0.243
|
|
|
|
|
chr1_+_100099353
|
0.243
|
NM_000645
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr14_+_19735334
|
0.242
|
NM_001005503
|
OR11G2
|
olfactory receptor, family 11, subfamily G, member 2
|
|
chr14_+_19513442
|
0.241
|
NM_001005486
|
OR4K15
|
olfactory receptor, family 4, subfamily K, member 15
|
|
chr17_-_16816113
|
0.240
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr5_-_180560535
|
0.235
|
NM_203297
|
TRIM7
|
tripartite motif containing 7
|
|
chr22_-_14829803
|
0.233
|
NM_001005239
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1
|
|
chr1_+_6537924
|
0.231
|
NM_138697
NM_177540
|
TAS1R1
|
taste receptor, type 1, member 1
|
|
chr9_-_99746923
|
0.231
|
NM_018437
|
HEMGN
|
hemogen
|
|
chr20_-_3635774
|
0.227
|
NM_023068
|
SIGLEC1
|
sialic acid binding Ig-like lectin 1, sialoadhesin
|
|
chr11_-_116168316
|
0.224
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr3_+_35697432
|
0.223
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr19_-_12858956
|
0.222
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr2_-_25320582
|
0.222
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr7_+_31693155
|
0.221
|
NM_001145123
NM_006658
|
C7orf16
|
chromosome 7 open reading frame 16
|
|
chr17_+_71975224
|
0.219
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr6_-_11490487
|
0.218
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_-_55130925
|
0.217
|
|
RTN4
|
reticulon 4
|
|
chr16_+_76613912
|
0.215
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr12_-_121753795
|
0.214
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr7_+_138695209
|
0.214
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr19_+_56452775
|
0.214
|
NM_173635
|
C19orf75
|
chromosome 19 open reading frame 75
|
|
chr22_-_16127520
|
0.211
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr11_-_71424614
|
0.211
|
|
|
|
|
chr12_-_53064565
|
0.209
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr17_+_35471971
|
0.207
|
NM_001190919
|
THRA
|
thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian)
|
|
chr1_+_213245507
|
0.207
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr3_+_52788976
|
0.207
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr10_-_121286034
|
0.206
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr3_+_46894239
|
0.204
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr22_+_34979062
|
0.202
|
NM_001136540
NM_001136541
NM_003661
NM_145343
|
APOL1
|
apolipoprotein L, 1
|
|
chr12_+_99274987
|
0.200
|
NM_001145288
NM_139319
|
SLC17A8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8
|
|
chr3_-_129668780
|
0.200
|
NM_153330
|
DNAJB8
|
DnaJ (Hsp40) homolog, subfamily B, member 8
|
|
chr11_+_5558682
|
0.198
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr12_+_27511009
|
0.197
|
NM_001145010
|
C12orf70
|
chromosome 12 open reading frame 70
|
|
chr7_+_94130752
|
0.196
|
|
PEG10
|
paternally expressed 10
|
|
chr19_-_56537189
|
0.195
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr1_-_199613406
|
0.194
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr19_-_11549484
|
0.194
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr2_-_55130963
|
0.194
|
|
RTN4
|
reticulon 4
|
|
chr3_+_142525744
|
0.193
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr11_-_56914668
|
0.192
|
NM_002728
|
PRG2
|
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein)
|
|
chr9_-_135435165
|
0.191
|
NM_001080515
|
FAM163B
|
family with sequence similarity 163, member B
|
|
chr11_-_16454493
|
0.190
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr6_-_27988152
|
0.190
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr11_-_614901
|
0.189
|
NM_001171968
NM_021924
NM_031264
|
CDHR5
|
cadherin-related family member 5
|
|
chr20_-_30795463
|
0.189
|
NM_001099339
NM_053041
|
COMMD7
|
COMM domain containing 7
|
|
chr2_-_65013070
|
0.186
|
|
LOC400958
|
hypothetical LOC400958
|
|
chr7_+_138695241
|
0.185
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr17_+_36642240
|
0.183
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr12_+_1608672
|
0.181
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr17_-_10362583
|
0.181
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr6_-_41811916
|
0.179
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr10_-_921694
|
0.178
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr1_+_158063278
|
0.177
|
|
SLAMF8
|
SLAM family member 8
|
|
chr12_-_2814410
|
0.176
|
NM_031474
|
NRIP2
|
nuclear receptor interacting protein 2
|
|
chr6_+_163068991
|
0.176
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr6_+_126282078
|
0.174
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr19_-_11549441
|
0.173
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr10_-_105427898
|
0.173
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_-_156800017
|
0.170
|
NM_001160325
|
OR6P1
|
olfactory receptor, family 6, subfamily P, member 1
|
|
chr22_+_43451600
|
0.167
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr2_-_10860093
|
0.166
|
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chr2_-_55131202
|
0.165
|
NM_020532
NM_153828
NM_207520
|
RTN4
|
reticulon 4
|
|
chr14_-_93512818
|
0.164
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chrX_-_55037235
|
0.163
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr5_-_158690047
|
0.162
|
NM_002187
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40)
|
|
chr6_-_30236667
|
0.161
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr10_-_98311813
|
0.160
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_+_242283129
|
0.160
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr19_-_38052481
|
0.160
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr2_+_187173176
|
0.157
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr6_-_163068749
|
0.155
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chrX_+_128741562
|
0.155
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr17_-_46479127
|
0.153
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr6_-_41811323
|
0.153
|
|
TFEB
|
transcription factor EB
|
|
chr11_-_2279642
|
0.153
|
NM_001142946
|
C11orf21
|
chromosome 11 open reading frame 21
|